peaKO
Finding transcription factor binding motifs using knockout controls
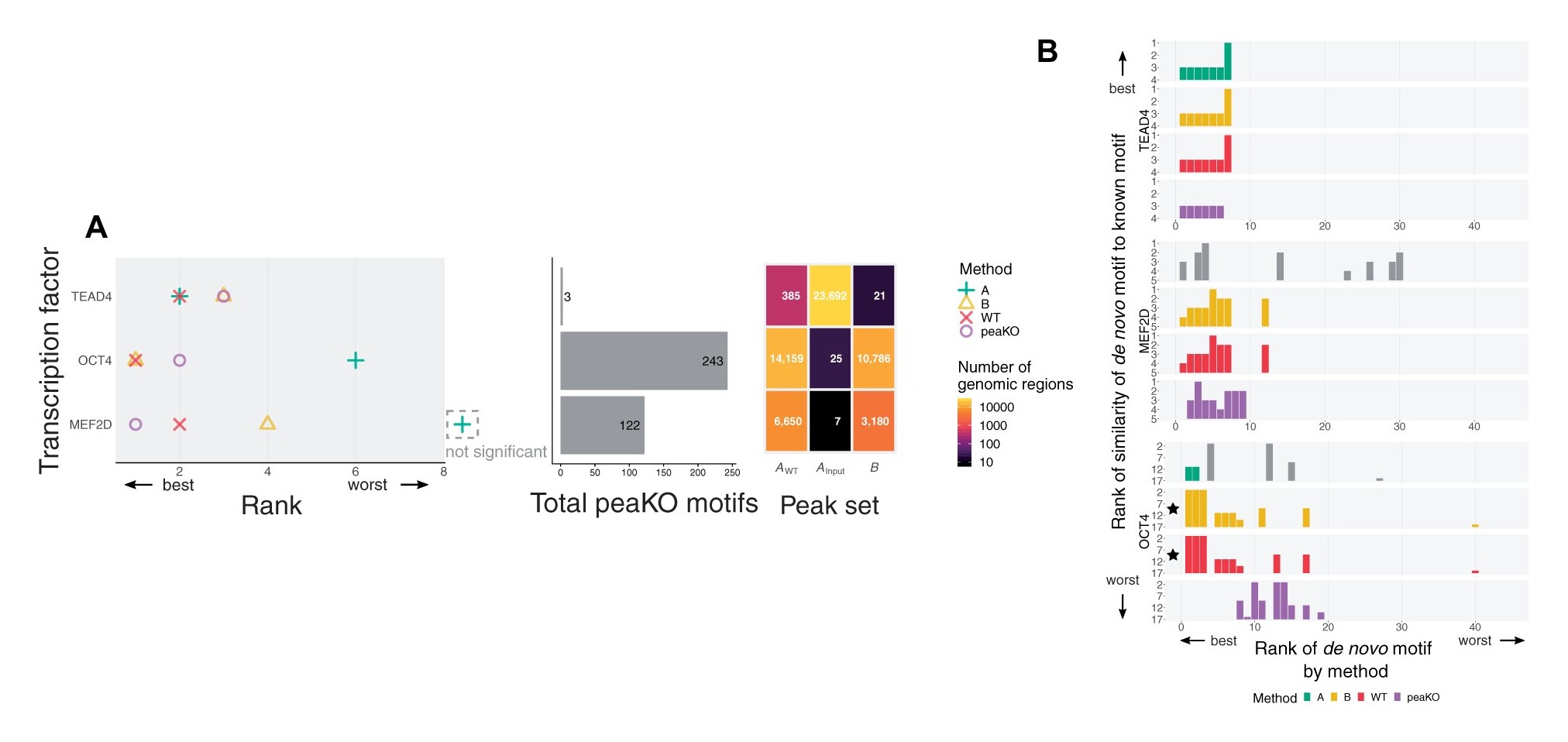
PeaKO is a computational tool to identify motifs relevant to ChIP-seq experiments by combining two differential analysis approaches. It often improves elucidation of the target motif over other methods and highlights the benefits of knockout controls. Click here for a detailed description page.
---
Denisko D, Viner C, Hoffman MM.
Motif elucidation in ChIP-seq datasets with a knockout control.
Bioinformatics Advances, 2023.
Available from: https://doi.org/10.1093/bioadv/vbad031.
---
I developed peaKO during my MSc at the University of Toronto. I used Snakemake to write the main workflow that ties together a series of Python scripts. It works with Slurm scheduling or just locally. The name “peaKO” (pee-koh) comes from “peak” and “KO” (knockout), but it’s also a reference to the name my grandma would have chosen for a hedgehog, as in “orange pekoe” :)
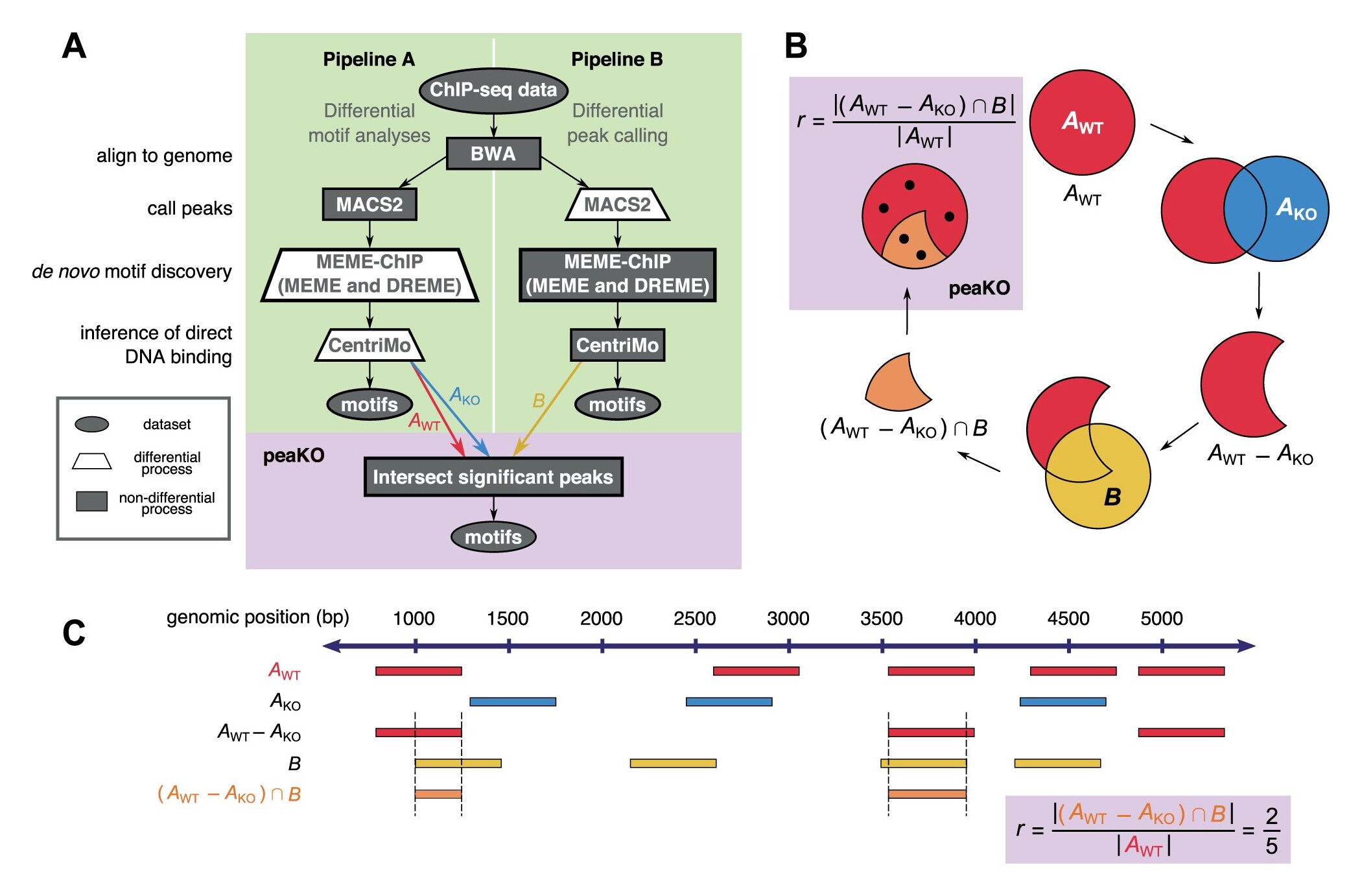
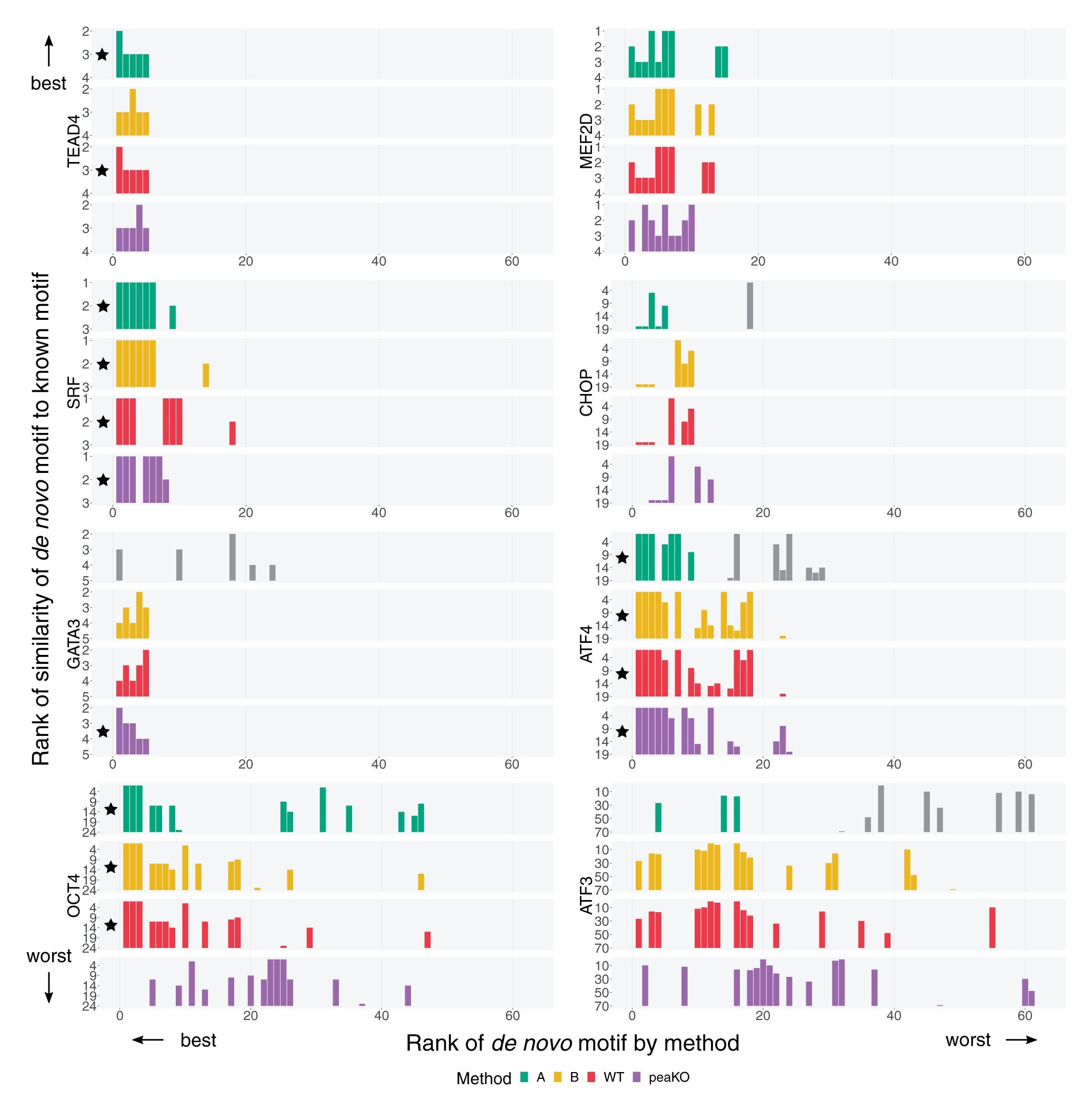
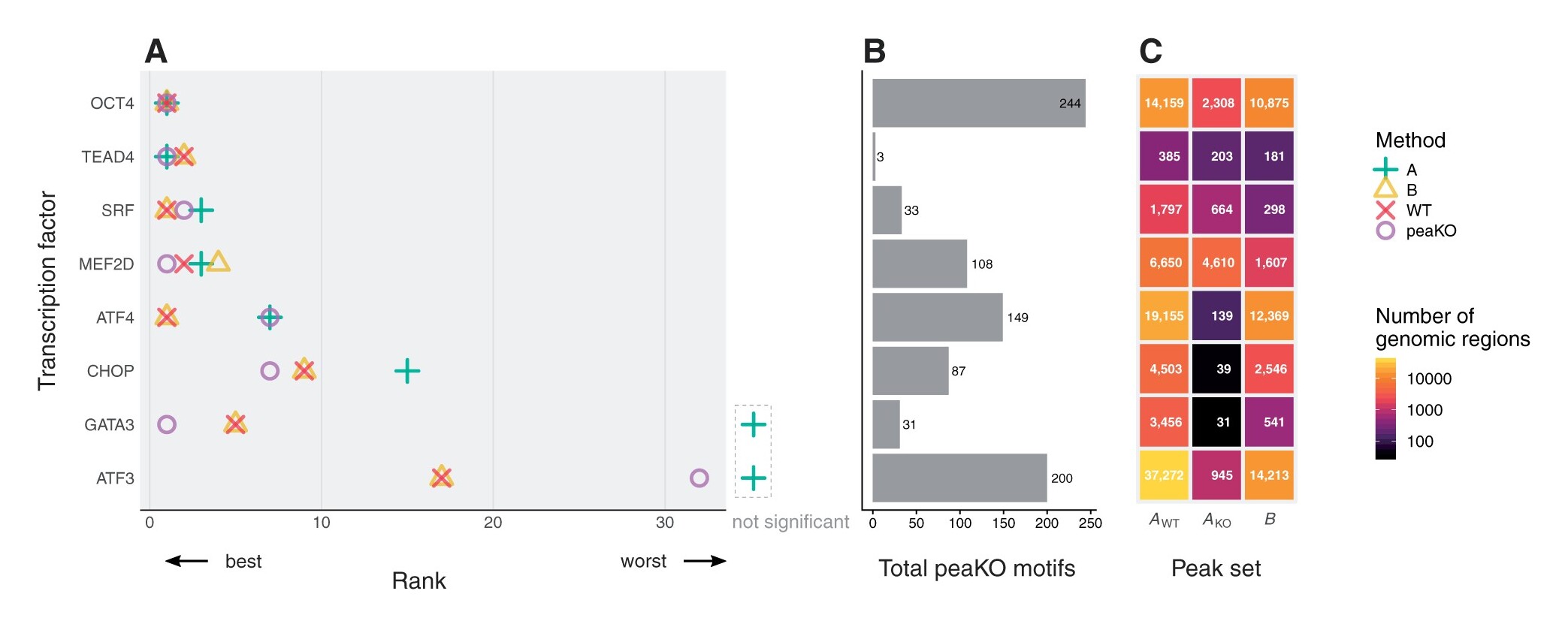
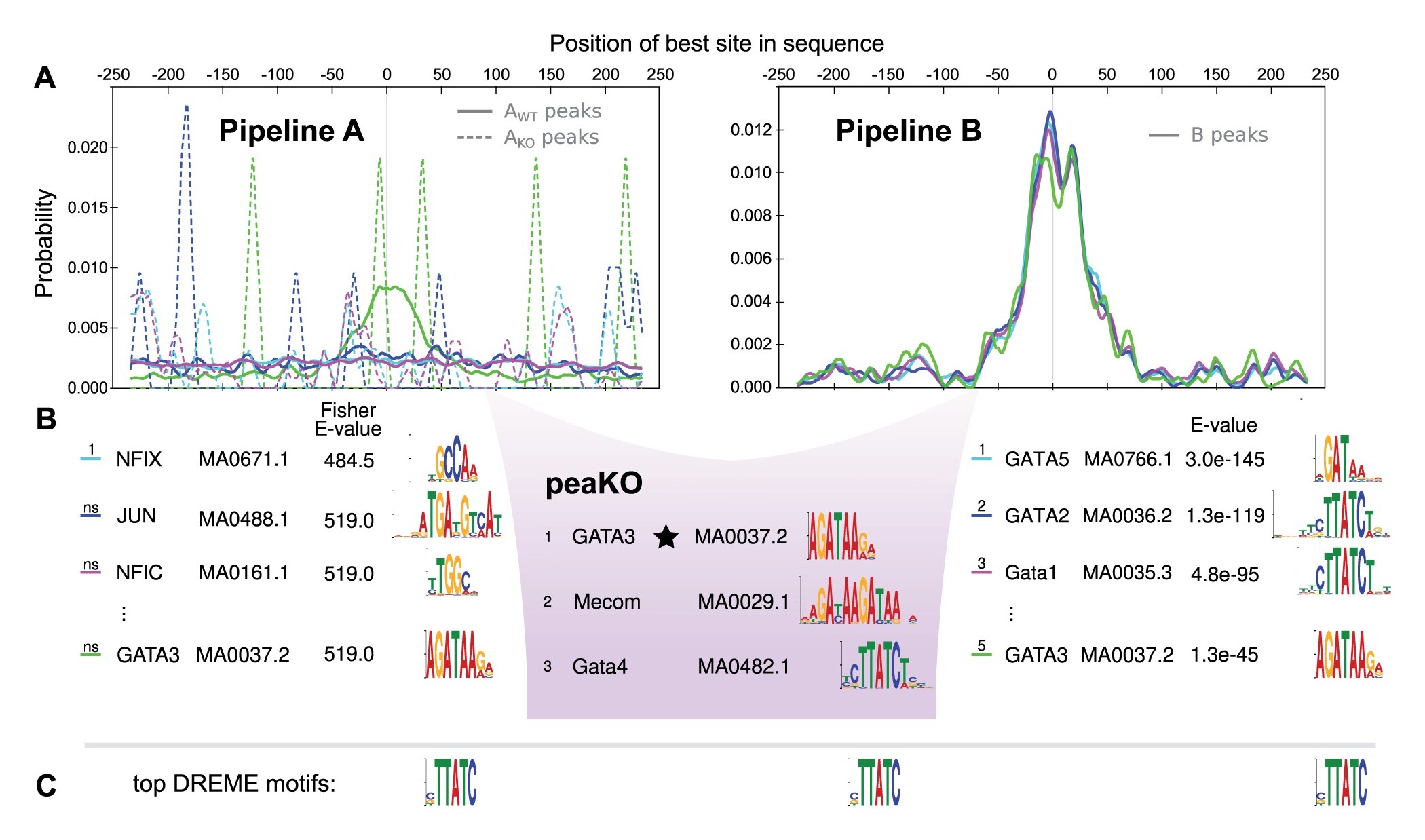
Longer description: PeaKO is a computational method for identifying transcription factor binding motifs from wild-type/knockout paired ChIP-seq datasets. PeaKO optimizes motif analyses by implementing a dual-pipeline approach. The first pipeline incorporates differential motif analysis, while the second incorporates differential peak calling. I combined these pipelines to select for motifs that both have consistent matches within peaks and fall within regions of significant read pileup. PeaKO computes a new metric based on the proportion of overlapping peaks between both pipelines, with overlaps interpreted as genuine binding events. PeaKO uses this metric to rank a collection of known or de novo motifs, where top-ranked motifs are thought to be relevant to the ChIP-seq experiment.
To install peaKO, follow the steps on GitHub:
conda create --name peako
conda activate peako # or source activate peako
conda install python=3.7
conda install -c anaconda beautifulsoup4=4.7 pandas
conda install -c bioconda -c conda-forge -c anaconda snakemake-minimal flake8 pathlib2 ipython twine
conda install -c bioconda pybedtools
python3 -m pip install peako
peako --help
# Usage: peako <outdir> <wt-bam> <ko-bam> <organism> <chr-sizes> <trf-masked-genome> <motif-database> [options]